About
I am an Assistant Professor in the Department of Statistics at George Mason University.
Prior to joining George Mason, I obtained a PhD in Statistics in 2020 from the University of British Columbia where I was advised by Dr. Gabriela Cohen Freue. Before that, I obtained a Master of Science in Statistics from the Vienna University of Technology under supervision by Prof. Peter Filzmoser.
My research agenda comprises methodological and computational aspects of robust estimation in high-dimensional problems as well as their application to Biomedical Sciences. I am working on statistical methods with reliable performance under the presence of adverse contamination anywhere in the numerous features of the data.
For regression problems, for instance, I work on estimators which are resilient to outliers in the response but also to unusual values in the (potentially) explanatory variables. If not handled appropriately, unusual values in the explanatory variables can have a much more detrimental affect on the analysis than outliers in the response alone.
Selected projects

Improving phenological modeling via participatory science
Can we guide the wisdom of the crowds to solve complex scientific problems?
We aim at improving phenological modeling by sourcing models from citizen scientists. The starting point for the project was our First International Cherry Blossom Prediction Competition, where we asked participants to predict the peak bloom date of cherry trees in four locations: Washington, D.C., Vancouver, BC, Kyoto, Japan, and Liestal-Weideli in Switzerland.
In the news
The 2021 edition of the prediction competition has been featured prominently in the news. For example, The Weather Network, CBC Radio, Public Radio's The World, the Vancouver Sun, and the Daily Hive reported on the competition and how it can inform future research.
Find out more about the competition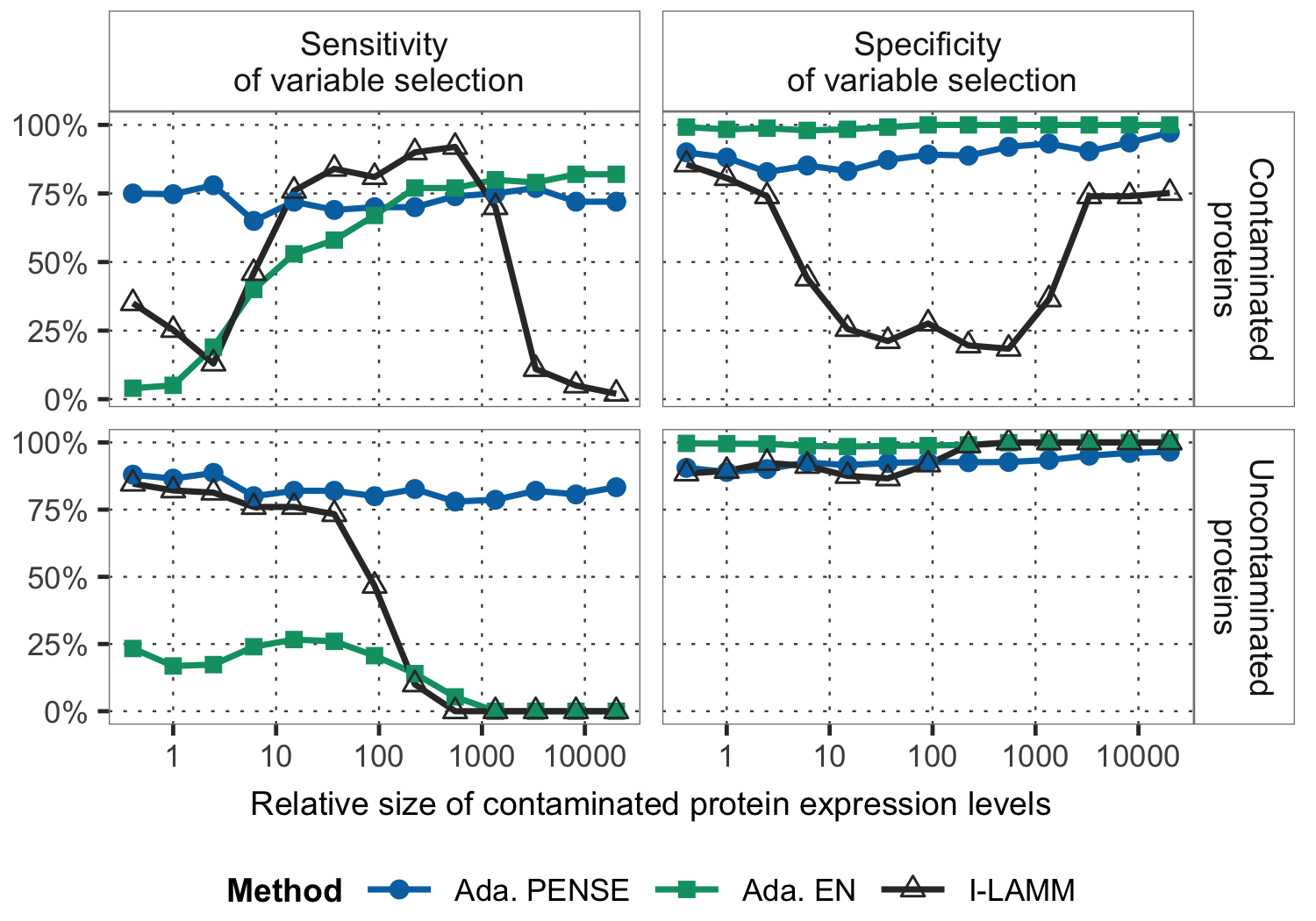
Robust estimation and variable selection in high-dimensional models
Outliers and contamination in data sets with many variables make common tasks like variable selection and parameter estimation a very daunting task. We are working on statistical methods and computational strategies for estimation, variable selection, and hyper-parameter selection to leverage as much information from the data set as possible, without being affected by misleading values.
Publications
Book Chapters
2022
Pre-prints
Refereed Journal Papers
2025
2024
2023
2019
2017
2013
Patents/Patent Applications
2024
Software
2024
2022
2021
2023
2019
Lab members

Yang Long
PhD Candidate at Mason
Yang Long is a member of my lab since 2021. His primary focus is on improving computation of robust regularized regression estimators by direct minimization of the non-convex objective function.

Siqi Wei
PhD Candidate at Mason
Siqi Wei joined my lab in 2021 and currently works on improving hyper-parameter selection for robust regularized regression estimators and general non-convex estimators.
Code
I am maintaining several stable R packages on CRAN and Bioconductor as well as a few experimental software tools available on my GitHub and GitLab pages.
pense
Implementation of penalized adaptive Elastic Net S/MM-Estimators of Regression
Robust penalized adaptive elastic net S- and MM-estimators for linear regression.
More info View on CRANexaminr
Create online exams from R markdown documents.
Write online exams as R markdown documents and publish them as shiny app. Allows for randomized exams, different question types (including R coding questions), and grading of submissions.
More infopyinit
Peña-Yohai Initial Estimator for Robust S-Regression
Fast and deterministic procedure to compute initial estimates for robust S-estimators of regression using as described in Peña-Yohai (1999).
View on Github View on CRANgaselect
Genetic algorithms for variable selection.
Multi-threaded genetic algorithms applicable to a wide range of variable selection methods, but particularly suited for Partial Least Squared Regression.
View on CRANcomplmrob
Robust Linear Regression with Compositional Covariates
Methods for robustly fitting regression models where the explanatory variables are compositional. Includes bootstrap methods for classical robust regression and compositional robust regression.
View on CRANnsoptim
Algorithms for non-smooth optimization
C++ template library, wrapped in an R package, providing modern and fast algorithms for optimizing non-smooth functions (e.g., L1 regularized objective functions).
View on GitLabPGCA
Link Protein Groups Created from MS/MS Data
Protein Group Code Algorithm (PGCA) is a computationally inexpensive algorithm to merge protein summaries from multiple experimental quantitative proteomics data.
View on BioconductorTeaching
George Mason University
- STAT 463 – Exploratory Data Analysis: Fall 2024, Fall 2023
- STAT 507 – Introduction to Statistical Data Science: Spring 2025
- STAT 634 – Case Studies in Data Analysis: Spring 2022, Spring 2021
- STAT 665 – Categorical Data Analysis: Fall 2023, Fall 2021
- STAT 778 – Statistical Computing: Spring 2025, Spring 2024, Spring 2023
University of British Columbia
- STAT 305 – Introduction to Statistical Inference: Spring 2020







